|
|
Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells
Acknowledgements References |
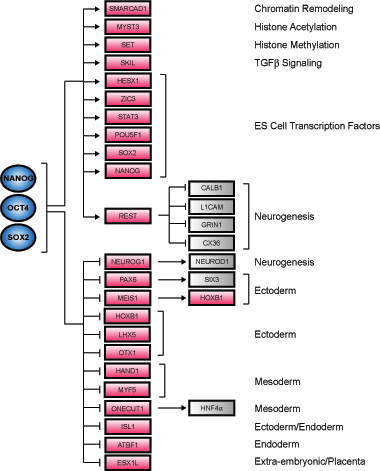
An initial model for ES cell transcriptional regulatory circuitry was constructed by identifying Oct4, Sox2, and Nanog target genes that encode transcription factors and chromatin regulators, and integrating knowledge of the functions of these downstream regulators in both human and mouse based on the available expression studies and literature (Figure 5). The model includes a set of active and a set of repressed target genes based on the extensive expression characterization of the 353 co-bound genes as described earlier. The active targets include genes encoding components of chromatin remodeling and histone modifying complexes (e.g. SMARCAD1, MYST3, and SET), which may have general roles in transcriptional regulation and genes encoding transcription factors (e.g. REST, SKIL, HESX1, STAT3) which themselves are known to regulate specific genes. |
| YOUNG
LAB
Whitehead Institute 9 Cambridge Center Cambridge, MA 02142 [T] 617.258.5218 [F] 617.258.0376 CONTACT US |