|
|
Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells
Acknowledgements References |
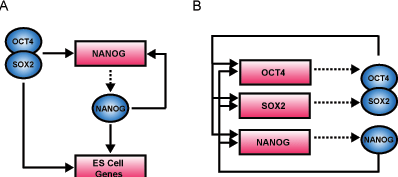
In order to identify regulatory network motifs associated with Oct4, Sox2 and Nanog, we assume that regulator binding to a gene implies regulatory control and have used algorithms that were previously devised to discover such regulatory circuits in yeast (Lee et al., 2002). The simplest units of commonly used transcriptional regulatory network architecture, or network motifs, provide specific regulatory capacities such as positive and negative feedback loops to control the levels of their components (Lee et al., 2002; Milo et al., 2002; Shen-Orr et al., 2002). |
| YOUNG
LAB
Whitehead Institute 9 Cambridge Center Cambridge, MA 02142 [T] 617.258.5218 [F] 617.258.0376 CONTACT US |